Problem Statement
Creating FAIR R&D data has become crucial as organizations adopt high-throughput methods and integrate AI/ML into their scientific workflows. Lab instrument manufacturers proliferate proprietary formats, sometimes further customized or configured by individual scientists, blocking achievement of FAIR data and resulting in an industry-wide interoperability crisis in labs where diverse instruments and software systems cannot readily speak to one another. This not only limits data utilization at scale but also burdens IT and automation teams. Moreover, companies typically address these issues independently, solving the same problems repeatedly across the industry—leading to wasted resources and diminished scientific productivity.
Idea Proposal and Value Proposition
We propose a community-driven open-source approach to address this challenge. By pooling resources, the community can develop software to transform instrument outputs into common data standards. Open-sourcing these converter codes enables continuous usage, updates, and enhancements, ensuring robust and evolving solutions. Although open-source initiatives require an initial investment, they have shown long-term sustainability and adaptability across various industries. Because pharma companies typically use a common set of instruments, pooling resources to address these instruments result in significant savings for each company, while ensuring the problem is solved in a manner that is future proofed. It also helps inform instrument manufacturers about what the industry values in terms of data standardization and accessibility.
In 2023, Benchling published Allotropy, an industry-first open-source project for converting lab instrument data into the Allotrope Simple Model (ASM). We chose ASM because it is:
- Developed and maintained by the Allotrope Foundation, a consortium of industry stakeholders, ensuring broad applicability.
- Open-source, making it freely available and eliminating lock-in risk as a vendor-neutral format
- Specified by instrument class, providing flexibility across diverse scientific contexts and data.
Since its launch, Allotropy has demonstrated the viability of this approach: Benchling has shipped support for 35 instruments under the MIT open-source license. Some customers are also starting to contribute feature enhancements to the library. To further scale this initiative, we propose that Pistoia Alliance leads an industry-funded consortium to accelerate the development, adoption, and maintenance of the Allotropy library. Pistoia members that contribute to this project can help define the initial set of target instruments and scientific use cases, thus maximizing the ROI for their investments.

Join the Allotropy Open-Source Initiative
Your skills can help create tools that make instrument data more consistent, reusable, and FAIR for the industry. Reach out to Project Manager, Christian Baber, to participate.
Targeted Outputs
We propose that the project is divided into 3 phases: Phase 0 defines the priority instruments and selects a vendor; phase 1 builds converter codes and deposits in Allotropy; phase 2 maintains and grows the library. Benchling will contribute resources [TBD] (e.g. software development, managing code review, conducting stakeholder interviews).
Phase 0: Identify the target instrument list. Conducting interviews with contributing members to identify a common set of 20-30 target instruments that also have corresponding Allotrope Simple Models. By focusing on instruments common to members, the project ensures each members’ investment also benefits others, thus multiplying the ROI.
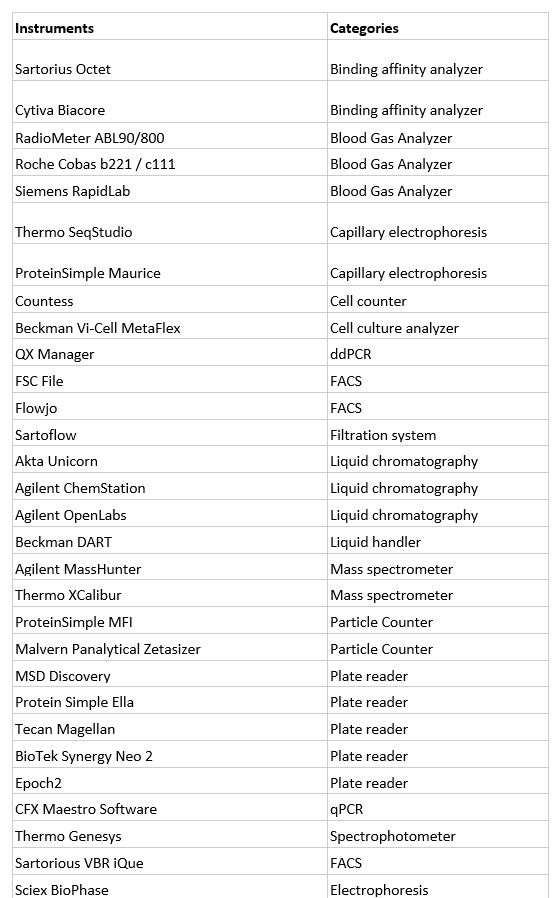
We think that 20-30 data parsers can address a significant number of use cases, since each parser can address multiple instrument models that share the same software system and that are commonly used for a scientific assay. For example, Other relevant information contains a list of the instruments that Benchling customers often request for integration. Together with the existing Allotropy parsers, we estimated that adding these additional 30 instrument parsers can already address ~ 70% typical scientific use cases.
In parallel, we will conduct a process to select a third-party vendor for the next phase. We expect Phase 0 to take 3-4 months. Benchling can contribute resources conducting this phase in collaboration with Pistoia Alliance.
Phase 1: Build the parsers. In this phase, Pistoia Alliance oversees the chosen vendor(s) to build the parser codes according to Allotrope Simple Model spec. We expect this phase to be conducted in collaboration with the Allotrope Foundation as additional features may need to be incorporated into the data models.
In our experience working with vendors such as Zifo, it typically takes 20-30 hrs to build a parser for a simple instrument and 40-60 hrs for a complex one. Hence for the 20-30 target instruments identified in phase 0, we estimate a total of 800~1200 hrs of work.
For this phase, Benchling can contribute resources to develop the parsers and to manage the Allotropy code review given our existing experience working with Allotrope foundation and Allotrope Simple Models. We expect this phase to take 6-9 months. We propose to cap the cost of this phase at [TBD, e.g. $150K].
Phase 2: Maintain and grow the library. As the industry adopts these parsers, Pistoia Alliance will oversee the maintenance and feature enhancements to the parsers. Based on the learnings from phase 0 and 1, we expect to build parsers for additional instruments. Finally, as instrument makers update their software, Pistoia Alliance is in a unique position to coordinate the industry effort of updating parsers. This phase will likely require a recurring annual budget from members. We will be able to have a clearer estimate of the cost after phase 0 and 1.
Critical Success Factors
Ongoing support and maintenance plan and funding. PA does not currently have resources or a platform to provide access to or maintain software.
Other Relevant Information
Common data generating lab instruments
Date Submitted: 11-Nov-2024
Author: Milton Yu
Idea Originators and Companies: Milton Yu (Benchling)
Strategic Priority: Delivering Data Driven Value

