Why is this important?
There is an enormous amount of unstructured, scientific research data and that is growing exponentially. As a result, researchers waste a lot of time trying to find documents, search data, understand it and interpret how to use it. Because the data is “un-FAIR”, researchers need to repeat experiments due to poor quality results, causing managers to make poor decisions.
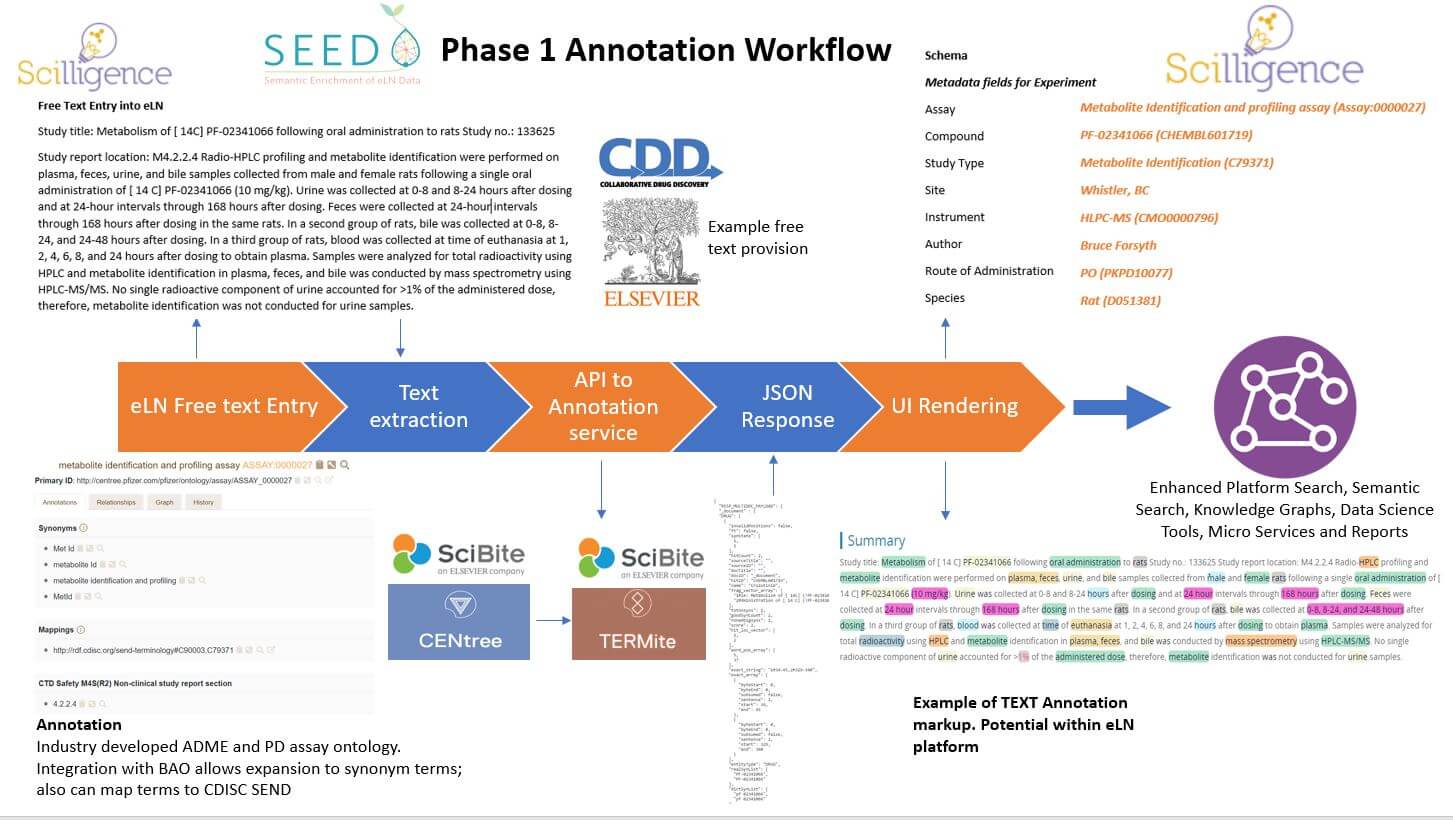
Semantic enrichment technology has advanced significantly in recent years and by applying this to text captured as part of an experiment, we will increase the value and use of the data. By using standardized terms with additional parameters to map relationships between terms, the data captured is enriched, more easily searched, and interoperable (FAIR).
This project objectives were to address a number of common issues:
- Volumes of unstructured eLN content are exponentially growing with the potential to extract knowledge
- Most eLN content is free text, unstructured information and thus challenging to identify common concepts (Named Entity Recognition NER), to effectively search and for extraction of deeper insights
- Retrospective data analysis and searching of generally unstructured content is challenging
- Data Workflows to/from eLNs can be relatively restricted in approach. Connectivity to Study
- Registration systems to enable Scientists to reuse such metadata through API to eLN are a must
- The availability of persistent identifiers challenges the capability of making data to be digitally discoverable and hinders aggregation of data inter-applications
- Valuable insights are hampered by unstructured content, preventing deeper data analysis and reducing competitive advantage
- When pharma companies license multiple eLN applications, a holistic approach and guidance for enabling consistent semantic enrichment across data in these applications is desirable
This will significantly increase researcher productivity by reducing rework (rerun experiments) and analysis time. High-quality results will provide improved insights and conclusions, enabling more effective decision-making.
Results
Project Completed – September 2023
The Journal of Biomedical Semantics has published a paper on our SEED project: “An extension of the BioAssay Ontology to include pharmacokinetic/pharmacodynamic terminology for the enrichment of scientific workflows”. The paper highlights the rich set of expertise that the project brought together and the work that was carried out in phase 1 of the project. You can read it here
https://jbiomedsem.biomedcentral.com/articles
Phase 2
Delivery of a new standard for Drug Safety terms and adds to Bioassay ontology, This will enable Assay/Study type ontology coverage for all of eCTD Module 4 of NDA (New Drug Application) submission.
- Extend Ontology work to include Drug Safety (3/4 months)
- Continue with Phase 1, including parameters leading to analysis e.g. knowledge graphs (2/3 month)
- Develop a workflow for structured data (e.g., Registration) (4/5 months)
- Scale-up across major ELN providers delivering an agnostic solution (8/9 months)
Phase 1
- A new standard assay Ontology (ADME PD) available to the community
- A working exemplar of ADME and PD workflow to semantically tag unstructured text
- An active community that supported Phase 1 and are sharing ideas for Phase II